Spatial regions
Spatial Regions
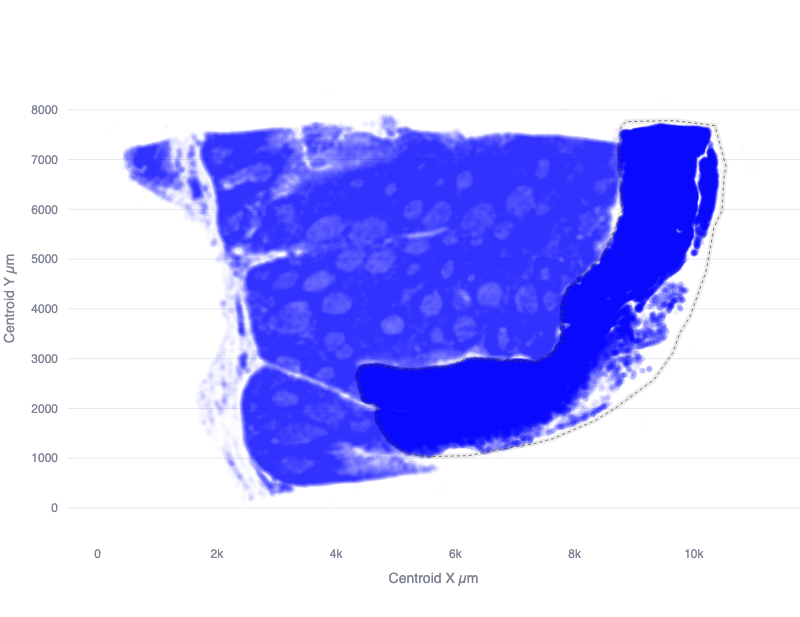
To analyze an experiment using spatial datasets, it is often necessary to define subregions of a particular dataset which should be compared against each other. One example of this is the use of tissue microarrays (TMA), where a collection of tissue cores are arranged in a grid for combined analysis. To analyze a dataset consisting of TMAs, it is necessary to define the region of each measurement (e.g. a Xenium run) which contains the specific TMAs of interest for a particular comparison.
The Quantitative Spatial Analysis (QSA) open source software project defines spatial regions using a simple JSON syntax which lists the X and Y coordinates of the vertices of a polygon that outline each region.
{
"dataset": {
"cirro_source": {
"domain": "organization.cirro.bio",
"project": "000000000-0000-0000-0000-000000000000",
"dataset": "00000000-0000-0000-0000-000000000000",
"path": "data/analysis-subfolder"
},
"type": "xenium"
},
"outline": [
{
"x": [
1671.0085411393027,
1497.2881668808081,
1005.0804398150743,
3431.375000292045,
2811.7723321034155
],
"xref": "x",
"y": [
4049.5208189474565,
3861.008928715988,
3528.3408871310435,
4265.75504597767
],
"yref": "y"
}
]
}
While it is possible to construct these coordinates manually, it is recommended that the QSA app be used for this purpose.