Single cell azimuth projection
Single-Cell Azimuth Projection
Single-cell sequencing provides an incredibly detailed description of the genes being expressed by individual cells from a complex tissue. However, it can sometimes be difficult to interpret this information in a coordinated way across multiple specimens, analysis batches, or sampling modalities. The Satija lab published a reference-based approach (Azimuth), which projects newly collected datasets into the multidimensional space established for a set of curated tissue-specific atlases. This approach can be used to quickly annotate cell types and align UMAP ordinations for new datasets to facilitate rapid comparison.
The Azimuth analysis can be run on single-cell gene expression datasets in Cirro, and will produce as an output an updated Seurat (h5seurat) or Scanpy (h5ad) object which can be used for further downstream analysis.
Azimuth Human Motor Cortex Reference Atlas:
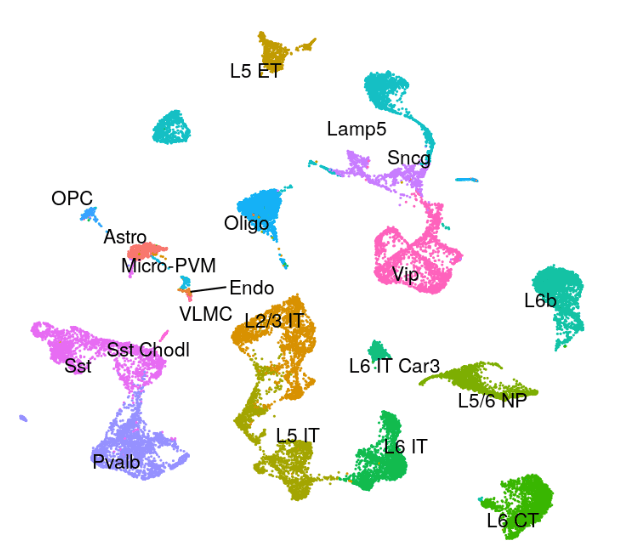
Azimuth References:
- Human Adipose -
adiposerefCells in Reference: 160,075 - Human Bone Marrow
bonemarrowrefCells in Reference: 297,627 - Human Fetal Development -
fetusrefCells in Reference: 377,456 - Human Heart -
heartrefCells in Reference: 656,509 - Human Kidney -
kidneyrefCells in Reference: 64,693 - Human Lung v2 -
lungrefCells in Reference: 584,944 - Human Motor Cortex -
humancortexrefNuclei in Reference: 76,533 - Mouse Motor Cortex -
mousecortexrefNuclei in Reference: 159,738 - Human Pancreas -
pancreasrefCells in Reference: 35,289 - Human PBMC -
pbmcrefCells in Reference: 161,764 - Human Tonsil -
tonsilrefCells in Reference: 263,299
Citation:
- Hao, Yuhan et al. “Integrated analysis of multimodal single-cell data.” Cell vol. 184,13 (2021): 3573-3587.e29. doi:10.1016/j.cell.2021.04.048
