Visualization Apps
For certain datasets, Cirro provides access to embedded applications to directly view and intepret the datasets in Cirro. For datasets that meet the requirements for you to use these apps, you will find them available as buttons at the top of the dataset's Overview page.
The currently available applications are:
Each button will include the app title and how many files are available to view for the dataset. Once you have opened the app, if more than one file is available to be viewed with the application, you will see a "Select a File" sidebar on the left side of the pop up which you can use to load different files. When you are done, you can collapse the sidebar and return to a fullscreen view of the app using the arrow at the top of the page.
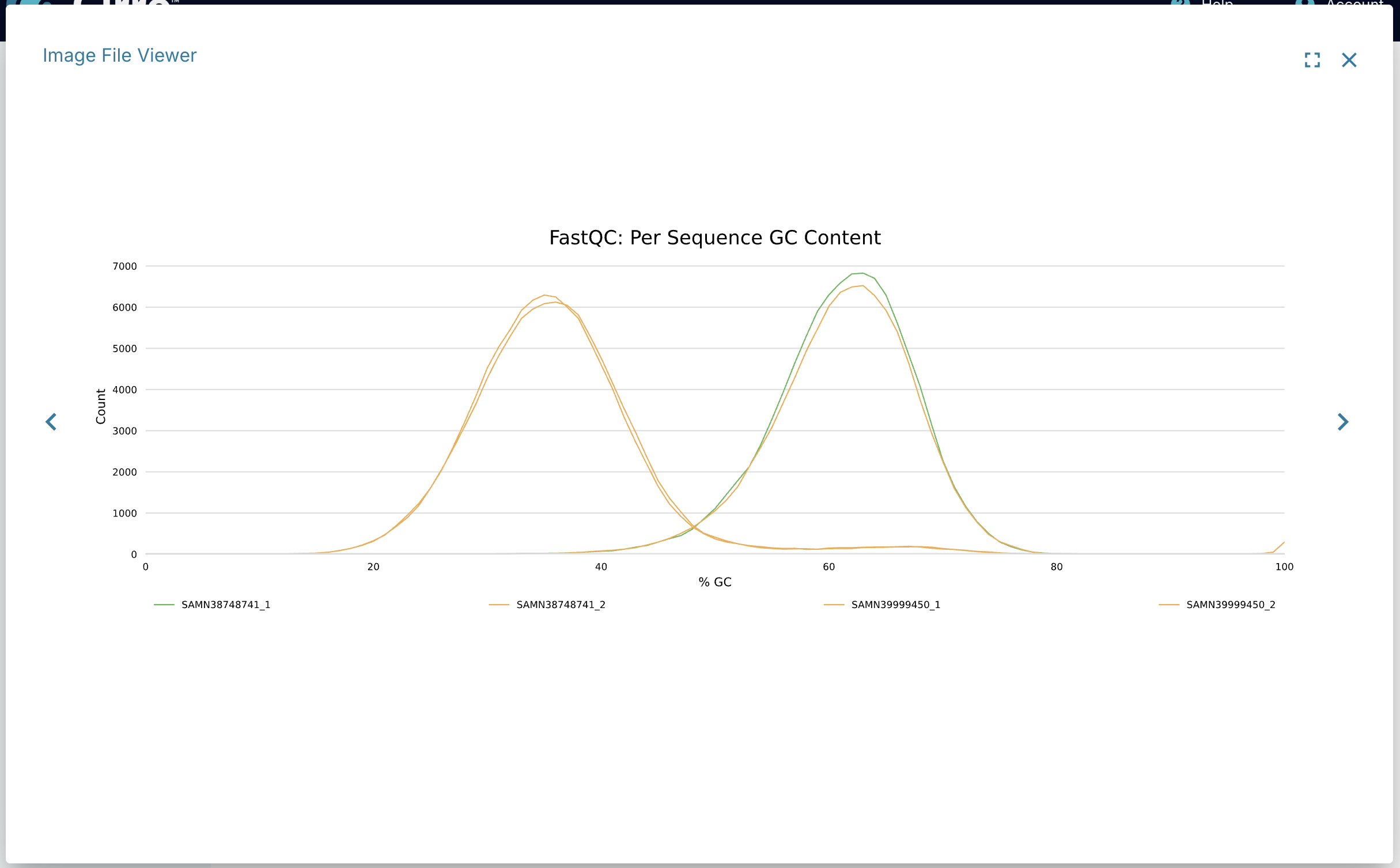
Image File Viewer
The Image File Viewer app allows you to view all image files in the dataset in one location. Any dataset that contains png, jpeg, or svg files will be able to view those files in the Image File Viewer.
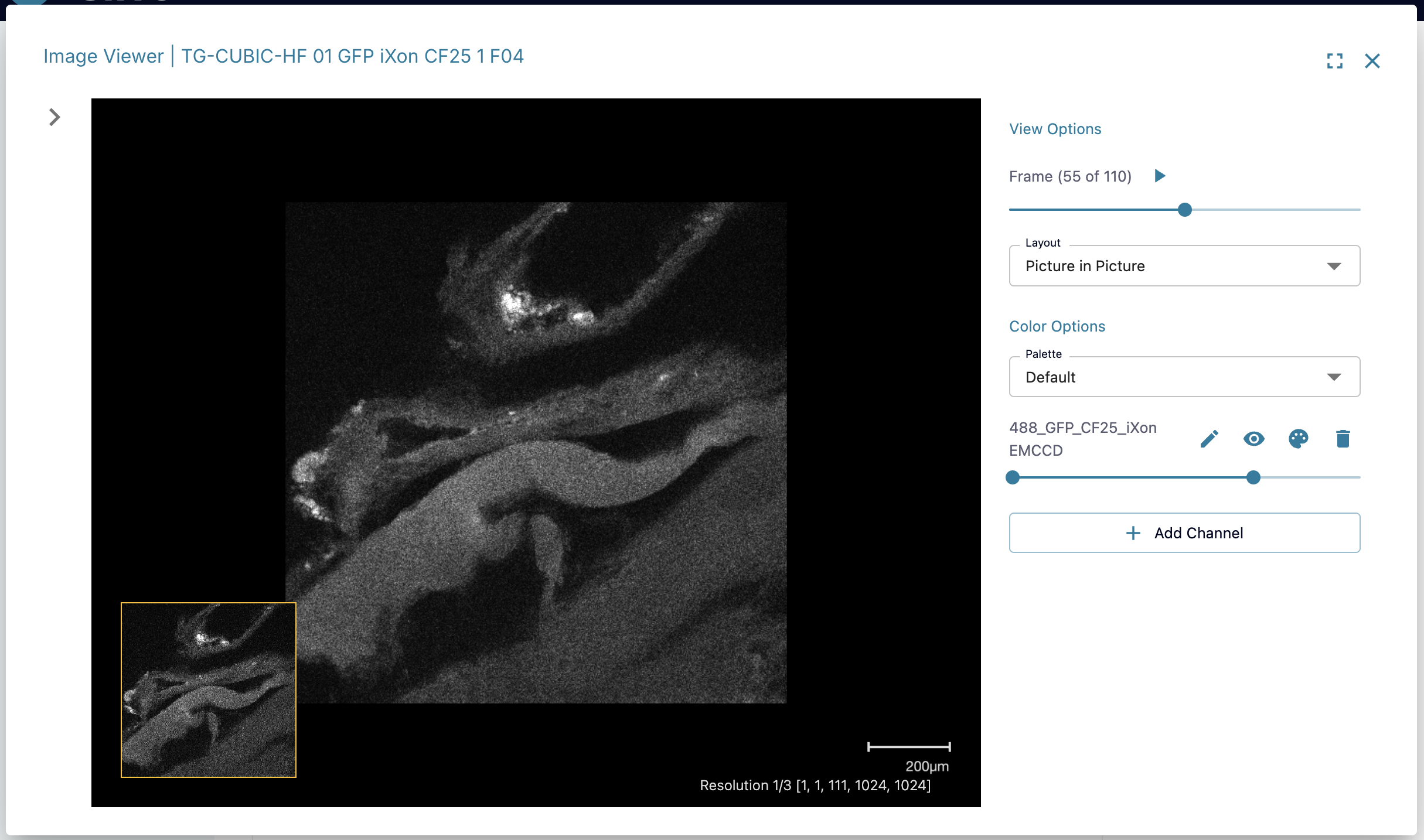
Microscopy Image Viewer
Input datasets uploaded with a dataset type of "Images" can use the Microscopy Image Viewer to visualize their files. This app can handle any of the following file types: .lif, .nd2, .ims, .tiff, .tif, .qptiff, .svs, .scn, .czi, .lsm, .ndpi. These file types will be converted by Cirro into OME-TIFF files which can be displayed using the Microscopy Image Viewer application.
Based on the contents of the original data files, you may have access to the following features when using the Image Viewer:
- Navigate between image frames
- Select between 2D and 3D layouts
- Set the image's color palette
- Add and adjust the different channels
- View any available file metadata
Learn more about OME-TIFF files.
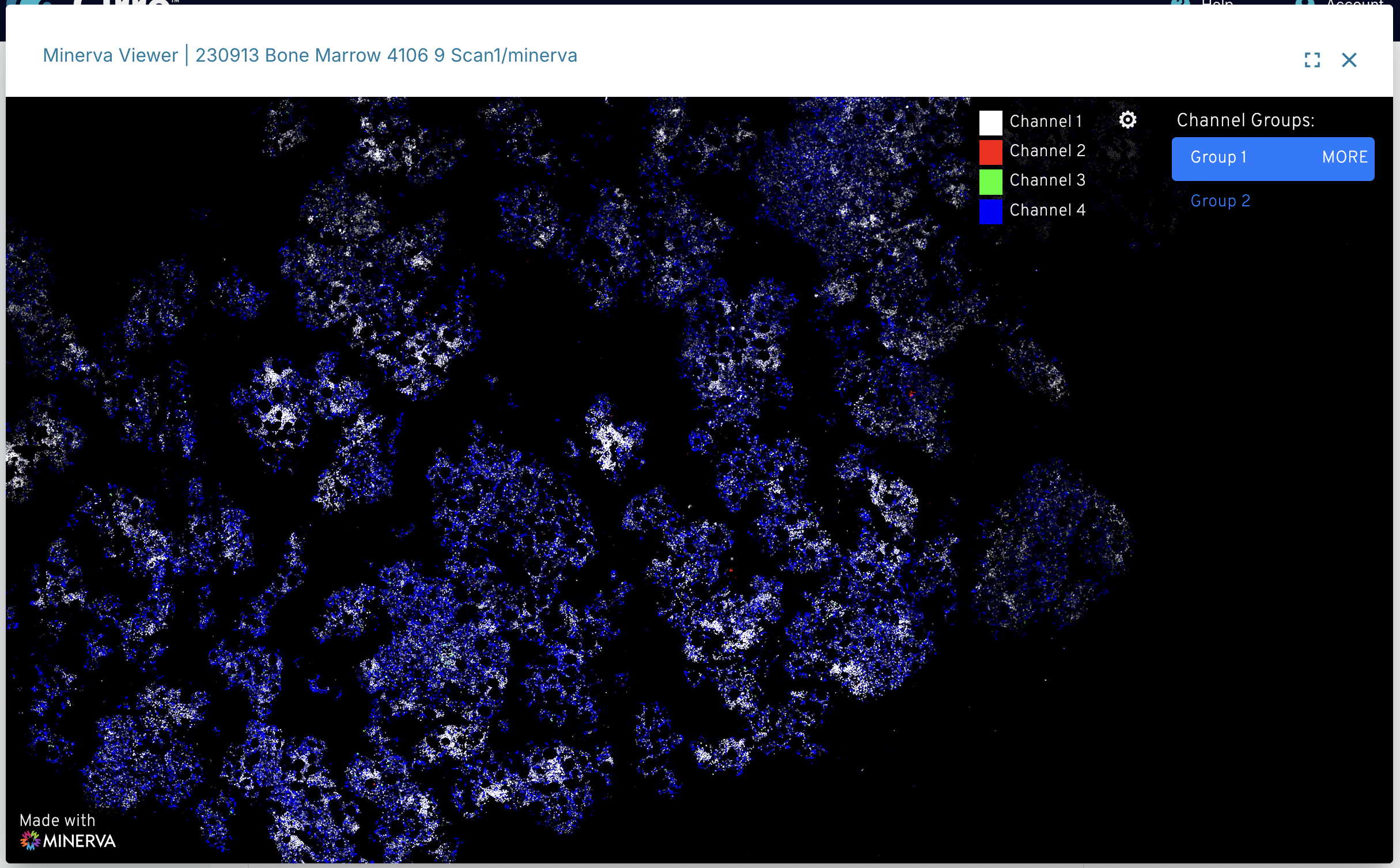
Minerva Viewer
Cirro has embedded the Minerva Story application to interact with large image files, like tissue atlases and digital pathology. Only datasets that have gone through a pipeline that is set up to create Minerva-compatible outputs, or was directly uploaded with these compatible files (using dataset type of "Minerva Story"), will be able to use the Minerva Viewer. Check any pipelines' documentation to see if Minerva outputs are enabled.
Learn more about using Minerva Story.
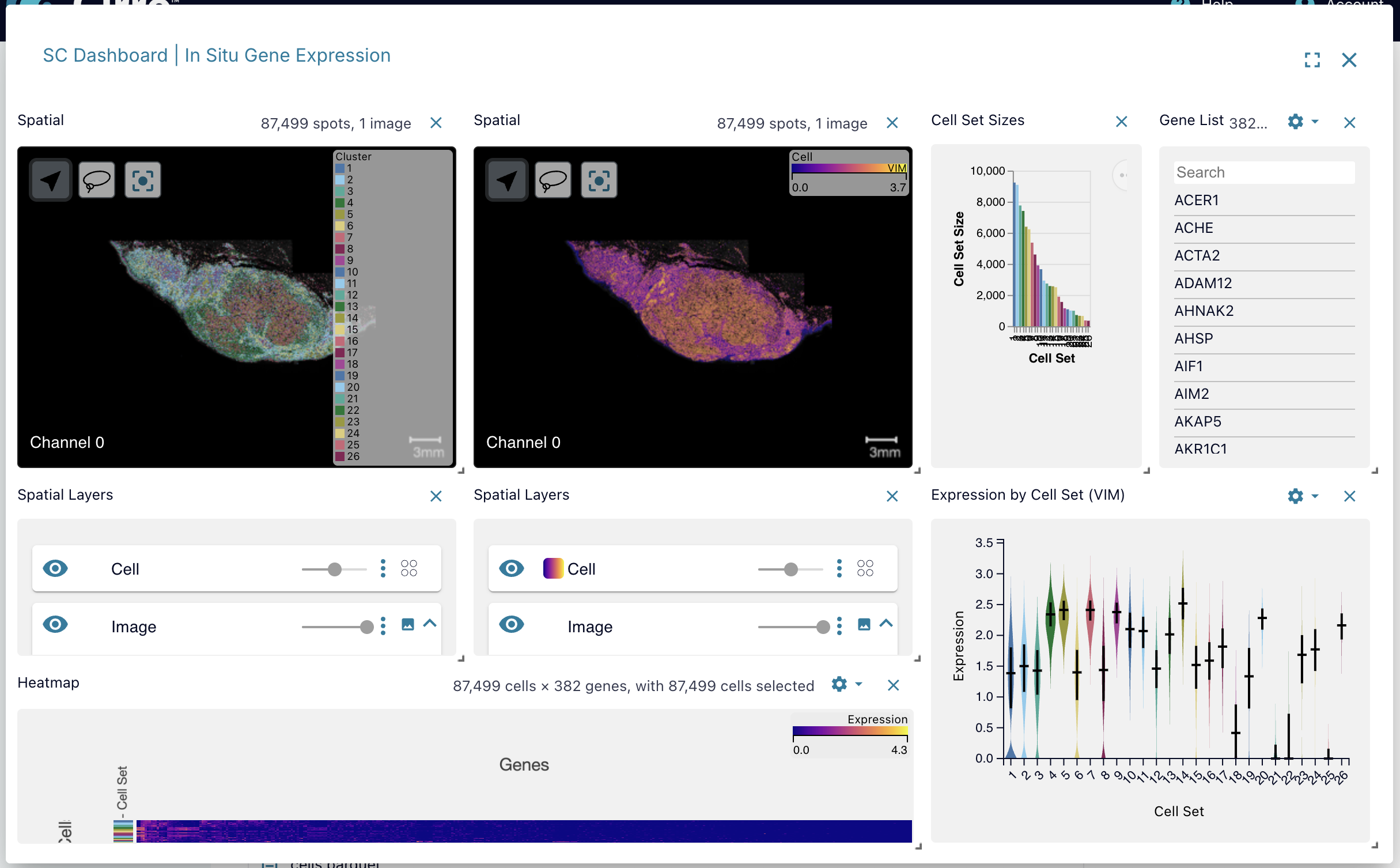
Single-Cell (SC) Dashboard
The results of certain single-cell gene expression analysis can be visualized directly in Cirro using an interactive display using Vitessce. Check the documentation for single-cell pipelines to learn if they support viewing outputs with this application. You can also upload your own spatial transcriptomics data using the "Vitessce Display" dataset type which requires a config.vt.json file and a Zarr file.
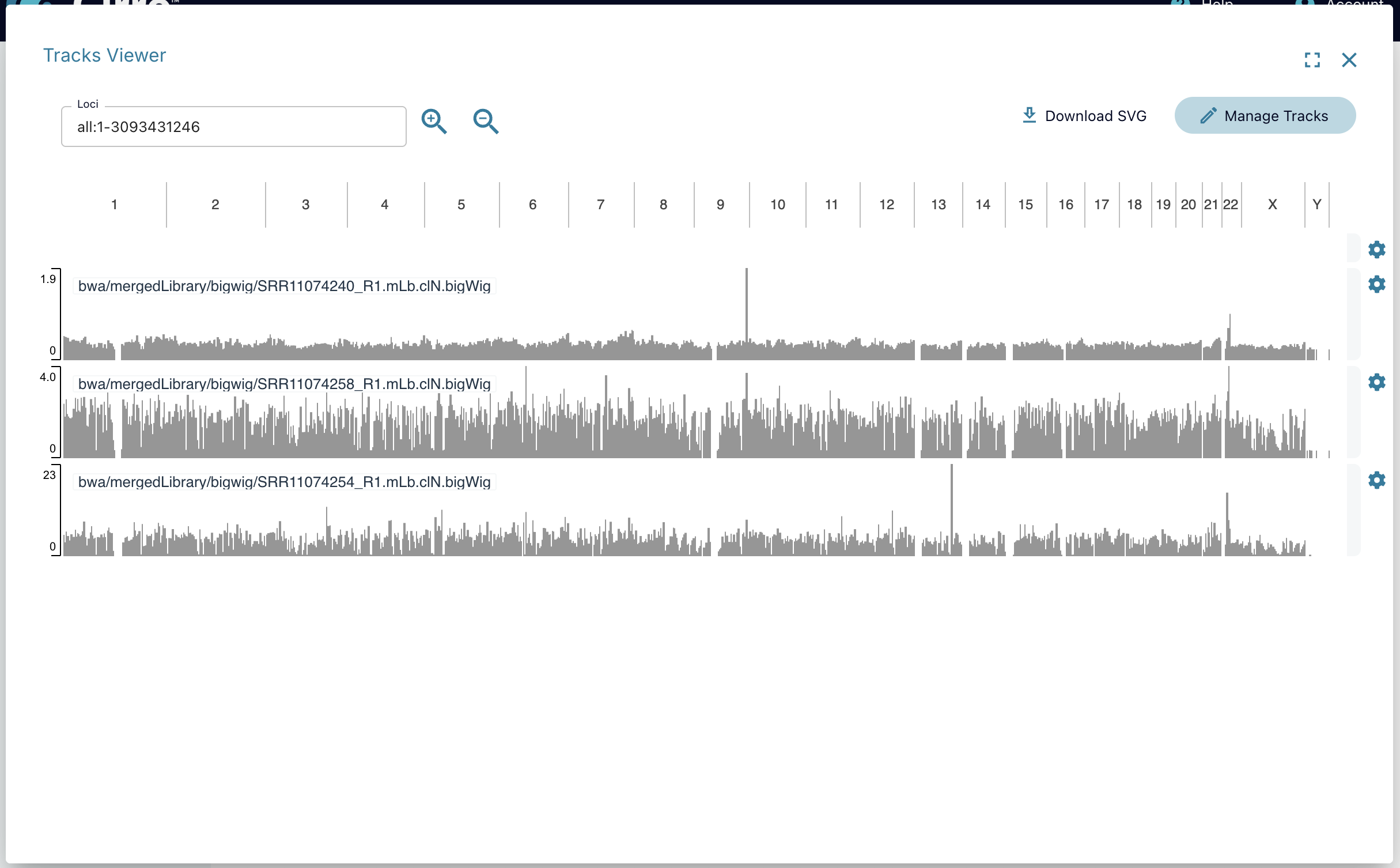
Tracks Viewer
Cirro has embedded the Integrative Genomics Viewer (IGV) to allow you to interact with your processed genomic data directly in Cirro. Datasets must have IGV-compatible files in order for the Tracks Viewer to show as an option,either by directly uploading the dataset or having gone through a pipeline that is set up to create those files. See the pipeline's documentation to check if it IGV outputs are enabled.
Learn more about using the IGV Track Viewer.